Achieving data maturity (3)
Part 3: From jupyter notebooks to CI/CD pipelines Still shipping notebooks?
Once you start learning Python, Jupyter Notebook becomes your buddy. That’s how most data professionals start. Others start in a .py env, which means they’re already working on production.
Notebooks were designed for exploration, demos, and teaching, but they become impractical for production pipelines, reproducibility, collaboration, and deployment.
Some companies still duct-tape them into “solutions,” and when their ML models collapse (like IKEA shelves), it becomes a problem.
If your production workflow still lives inside .ipynb files, you are not building yet and will still be considered an entry-level data person. (whatever that means).
1. Version control in notebooks is torture
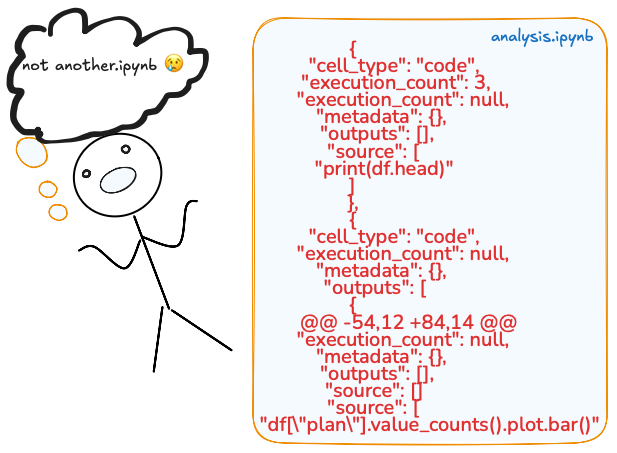
.ipynb files are not human-readable code files. They are like JSON blobs that store your code and metadata, like cell execution counts or output images. And because of this, -diffs are unreadable.
When you add a notebook to Git and try to review the changes later, Git displays a diff (the difference between 2 versions).
For a .py file that’s clean, but for a .ipynb file, you may be scrolling through lines of JSON completely blind.
The 1st one says little about what logic changed. You see cell numbers and output text blobs, metadata, and various other outputs, but the actual code that changed is buried within JSON fields. This makes PR a pain.
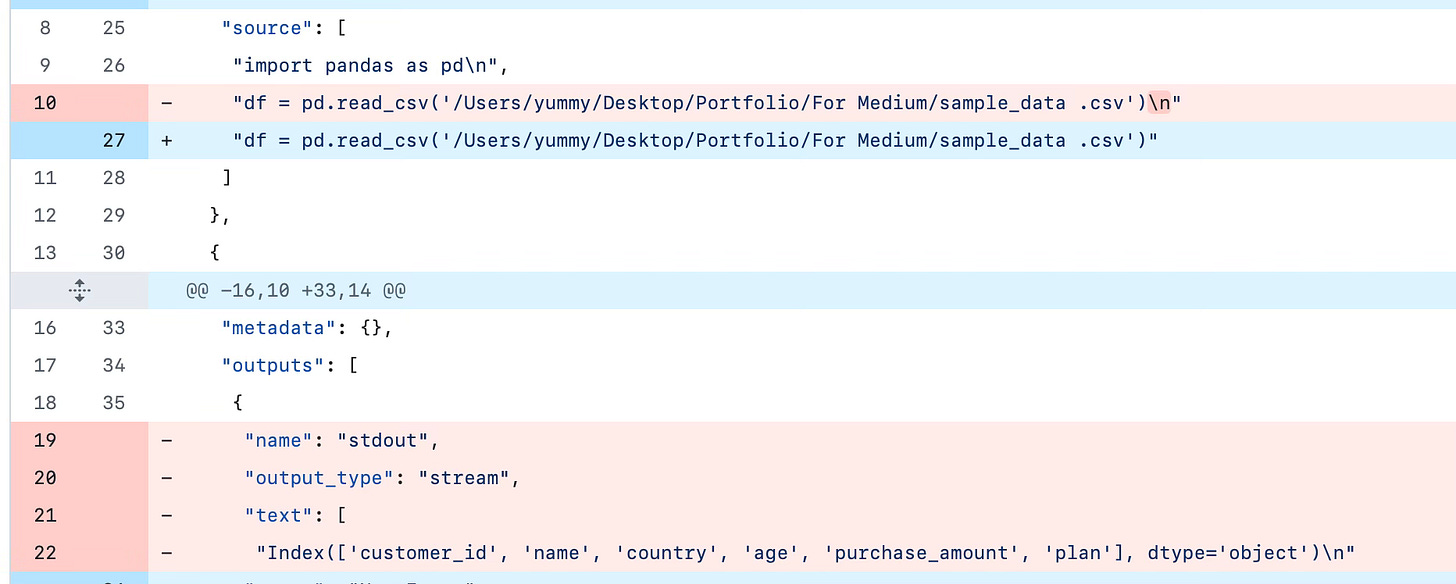
If you re-run the cells, say in a different order, add a plot, or get error messages, the JSON blows up:
ALL I DID WAS CHANGE “USA” TO “UK”. In a script, you can see the actual changes in the code in the git diff.
If someone reviews your commits and sees this, it’s already a long day.
- “execution_count”: 7,
+ “execution_count”: 8,
- “outputs”: [
- {
- “output_type”: “stream”,
- “text”: [
- “15\n”
- ]
- }
- ],
+ “outputs”: [
+ {
+ “output_type”: “stream”,
+ “text”: [
+ “104\n”
+ ]
+ }
+ ],2. Reproducibility
When running a notebook, execution matters, and notebooks don’t enforce it. i.e, If you restart a notebook, you must run cell 5 before cell 9.
import pandas as pd# 2
df = pd.read_csv(”customers.csv”)
print(df.head())# 3
df = df[df[”country”] == “US”]
df = df[df[”age”] > 30]
df = df[df[”purchase_amount”] > 100]# 4
summary = df.groupby(”plan”).size()
print(summary)Problem 1: double filtering: If you accidentally rerun cell 3, you’re filtering an already filtered df. Count drops again, and it does this silently. No error warning.
df = df[df[“country”] == “US”]
df = df[df[“age”] > 30]Problem 2: Out-of-order execution: if someone runs cell 4 before cell 2, you get a name error: NameError: name ‘df’ is not defined.
Even worse, if cell 2 were a different dataset in an earlier session, it would still produce results with the wrong data. If you run cell 4 before cell 5, the notebook will “remember” the last state and reproduce further results based on the old df.
That’s why 2 people may be working in the same notebook but have different results or inconsistencies in metrics. The notebook silently carried the state across the cells. Execution is stateful, not linear.
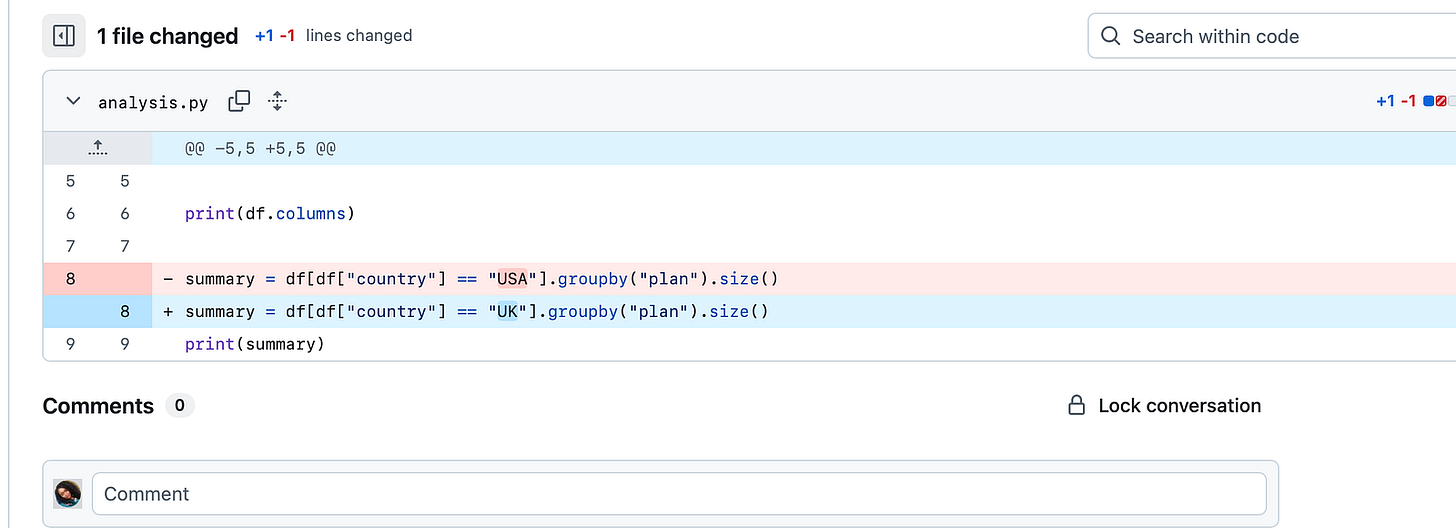
With a python pipeline.py, the execution order is always from top to bottom.
import pandas as pd
df = pd.read_csv(”customers.csv”)
df = df[df[”country”] == “US”]
df = df[df[”age”] > 30]
summary = df.groupby(”plan”).size()
print(summary)So if reproducibility matters to you, notebooks are not the best option, especially when you’re collaborating or at the production level. Notebook remembers state, scripts enforce order.
3. Testing & CI/CD gap
One major difference between playing around with data and working in a real-life data team is testing.
Simply put, it’s writing small checks that make sure your code does what you expect and functions don’t break when someone changes an input, or your model still works if there’s a new dataset.
In modern data teams, testing goes beyond your laptop, and that’s where CI/CD comes in. I’m not a data engineer, but I’m familiar with the basics of testing and building pipelines.
CI — continuous integration (every time you push code to GitHub)
CD — continuous deployment (if your tests pass, your code can be packaged and deployed, without manually taking files from one place to another).
So, how do you run a pytest on a notebook or integrate notebooks into a CI pipeline?
Short answer: you can’t. Tests require scripts (.py files) that can run in headless environments (no manual execution or hidden states). This can be done easily with a .py. With notebooks, you may need hacks likenbval, or you still have to convert to a .py first, so why not start with that in the first place?
Advanced teams work with .py files:
Using the analysis.py earlier, if you write a function, e.g
import pandas as pd
def summarize_by_country_plan(df, country):
return df[df[”country”] == country].groupby(”plan”).size()Next, write a test for it and call it tests/test_analysis.py:
##### tests/test_analysis.py
import pandas as pd
from analysis import summarize_by_country_plan
def test_summary_counts():
data = {
“country”: [”US”, “US”, “UK”],
“plan”: [”basic”, “premium”, “basic”]
}
df = pd.DataFrame(data)
result = summarize_by_country_plan(df, “US”)
assert result[”basic”] == 1
assert result[”premium”] == 1Run pytest locally:
pytestThe test passes when the code works. If someone changes the function and breaks the logic, the test will fail. If we drop this into a github action workflow:
# .github/workflows/test.yml
name: Run tests
on: [push]
jobs:
test:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v3
- name: Set up Python
uses: actions/setup-python@v4
with:
python-version: ‘3.10’
- name: Install dependencies
run: pip install -r requirements.txt
- name: Run tests
run: pytest tests/In this snippet, every time you push to github:
Python is installed
dependencies are pulled in
Every PR runs tests automatically, so if something breaks, you quickly know.
If you try to set that up for .ipynb , you’re locked out of this workflow. You’ll either
Run tests inconsistently (sometimes in CI, sometimes locally)
Or worse, manually run all cells before every merge.
That’s why advanced teams rarely stop at notebooks; they refactor code into .py modules, write tests, and let the CI/CD pipeline handle it.
Data maturity is the shift from exploration → reproducibility → scalability, and notebooks keep you at exploration.
Jupyter is useful, but data maturity means using something others can collaborate on.
Notebooks are for exploration; pipelines are for production. Data maturity is knowing the difference.
In the next part, I’ll unpack how this affects deployment, environment, and dependency chaos.
Till next time, see you in Part 4.
Amy
Enjoyed this? Read, comment, and subscribe for more.
Be data-informed, data-driven, but not data-obsessed
🔗 Biz and whimsy: https://linktr.ee/amyusifoh
🔗 Connect with me on LinkedIn and GitHub for more analytics.
🔗 Get the free design toolkit: dashboard design toolkit
Data analyst ⬩ Spreadsheet advocate ⬩ Freelancer ⬩ Turning data into useful insights
Read Part 1 here: Achieving data maturity (PART 1)






The great thing about Databricks is that notebooks are .py by default. Which sets it apart from other products like Snowflake. What that means is you can use and build tests in your CI/CD pipelines without having to hack it.
I’m a Databricks fanboy believe it or not had to drop my product knowledge 😂
Thanks for sharing. I think when people take a free Python intro workshop, the instructor usually uses notebooks, so I think that’s why people keep using them. Nostalgia?? lol